What is NGC?
High Throughput Sequencing (HTS) technologies produce enormous amounts
of digital data that have to be processed, transferred, stored and archived
which includes "raw" sequencing data as well as an even larger number of intermediate
and final result files that are produced by pipelines of data analysis and manipulation tools.
This motivates the development of data compression algorithms specialized for these data.
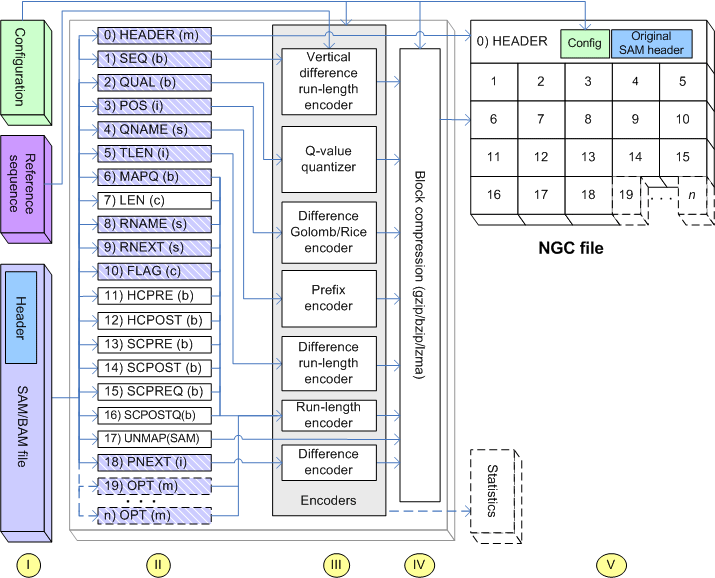
NGC is a compressor for aligned HTS sequencing data that enables the complete lossless and
lossy compression of mapped alignment data stored in SAM/BAM files.
NGC features
- NGC supports the efficient lossless compresssion of SAM/BAM files (space savings of 33-66%).
There are only minor differences between an original and an decompressed file:
- The order of the optional SAM tags is not preserved so they may appear in different order.
- The NM and the MD tags are automatically calculated from the data and may thus differ slightly due to SAM specification ambiguities.
- NGC supports lossy compression of per-base quality values in order to reach low compression ratios (save spacings up to 98%).
- Users may define different (non-)uniform quantization schemes for q-values of different categories.
- Users may specify an own quantization schema for unmapped reads
- Users may truncate read names and/or optional tags from a compressed file to save more disc space.
- Users may preserve q-values in special columns (e.g., explicitly listed ones in a VCF file or multi-allelic columns in low-coverage regions).
- Users may further quantize q-values in reads with low mapping quality.
- NGC supports multiple block compression methods (gzip/bzip2/lzma) that differ in speed and compression efficiency.
- NGC was implemented in Java and should run on all Java 1.7-enabled platforms.